Dynamic Estimation Objectives
The dynamic estimation objective function is a mathematical statement that is minimized or maximized to find a best solution among all possible feasible solutions. The form of this objective function is critical to give desirable solutions for model predictions but also for other applications that use the output of a dynamic estimation application. Two common objective functions are shown below as squared error and l1-norm forms1.
Squared Error Objective
The squared error objective is the most common form, used extensively in the literature. It is also the basis for the derivation of the Kalman filter and other well known estimators.
$$\min_{x,y,p} \Phi = \left(y_x-y\right)^T W_x \left(y_x-y\right) + \left(y-\hat y\right)^T W_m \left(y-\hat y\right) + \Delta p^T W_{\Delta p} \Delta p$$
$$\mathrm{subject\;\;to}$$
$$0 = f\left(\frac{dx}{dt},x,y,p\right)$$
$$0 \le g\left(\frac{dx}{dt},x,y,p\right)$$
l1-norm Objective
The l1-norm objective is like an absolute value objective but also includes a dead-band to reject measurement error and stabilize the parameter estimates.
$$\min_{x,y,p} \Phi = w_x^T \left(e_U+e_L\right) + w_m^T \left(c_U+c_L\right) + w_{\Delta p}^T \left(\Delta p_U+\Delta p_L\right)$$
$$\mathrm{subject\;\;to}$$
$$0 = f\left(\frac{dx}{dt},x,y,p\right)$$
$$0 \le g\left(\frac{dx}{dt},x,y,p\right)$$
$$e_U \ge y - y_x - \frac{db}{2}$$
$$e_L \ge y_x - y - \frac{db}{2}$$
$$c_U \ge y - \hat y$$
$$c_L \ge \hat y - y$$
$$\Delta p_U \ge p_i - p_{i-1}$$
$$\Delta p_L \ge p_{i-1} - p_i$$
$$e_U, e_L, c_U, c_L, \Delta p_U, \Delta p_L \ge 0$$
The l1-norm objective is like an absolute value of the error but posed in a way to have continuous first and second derivatives. The addition of slack variables enables an efficient formulation (only linear constraints) that is also convex (local optimum is the global optimum). A unique aspect of the following l1-norm objective is the addition of a dead-band or region around the measurements where there is no penalty. It is only when the model predictions are outside of this dead-band that the optimizer makes changes to the parameters to correct the model.
Nomenclature
There are many symbols used in the definition of the different objective function forms. Below is a nomenclature table that gives a description of each variable and the role in the objective expression.
$$\Phi=\mathrm{Objective\,Function}$$
$$y_x=\mathrm{measurements}$$
$$y=\mathrm{model\,predictions}$$
$$\hat y=\mathrm{prior\,model\,values}$$
$$w_x, W_x=\mathrm{measurement\,deviation\,penalty\,(WMEAS)}$$
$$w_m, W_m=\mathrm{prior\,prediction\,deviation\,penalty\,(WMODEL)}$$
$$w_{\Delta p},W_{\Delta p}=\mathrm{parameter\,movement\,penalty\,(DCOST)}$$
$$db=\mathrm{dead-band\,for\,noise\,rejection}$$
$$x,y,p=\mathrm{states,\,outputs,\,parameters}$$
$$\Delta p=\mathrm{parameter\,change}$$
$$f,g=\mathrm{equality\,and\,inequality\,constraints}$$
$$e_U,e_L=\mathrm{upper\,and\,lower\,error\,outside\,dead-band}$$
$$c_U,c_L=\mathrm{upper\,and\,lower\,deviation\,from\,prior\,model\,prediction}$$
$$\Delta p_U,\Delta p_L=\mathrm{upper\,and\,lower\,parameter\,change}$$
The following example problem is a demonstration of the two different objective forms and some of the configuration to achieve optimal estimator performance.
Exercise
Objective: Estimate parameters of a highly nonlinear system. Use an initialization strategy to find a suitable approximation for the parameter estimation. Create a MATLAB or Python script to simulate and display the results. Estimated Time: 2 hours
The spread of HIV in a patient is approximated with balance equations on (H)ealthy, (I)nfected, and (V)irus population counts2. Additional information on the HIV model is at the Process Dynamic and Control Course.
Initial Conditions H = healthy cells = 1,000,000 I = infected cells = 0 V = virus = 100 LV = log virus = 2 Equations dH/dt = kr1 - kr2 H - kr3 H V dI/dt = kr3 H V - kr4 I dV/dt = -kr3 H V - kr5 V + kr6 I LV = log10(V)
There are six parameters (kr1..6) in the model that provide the rates of cell death, infection spread, virus replication, and other processes that determine the spread of HIV in the body.
Parameters kr1 = new healthy cells kr2 = death rate of healthy cells kr3 = healthy cells converting to infected cells kr4 = death rate of infected cells kr5 = death rate of virus kr6 = production of virus by infected cells
The following data is provided from a virus count over the course of 15 years. Note that the virus count information is reported in log scale.
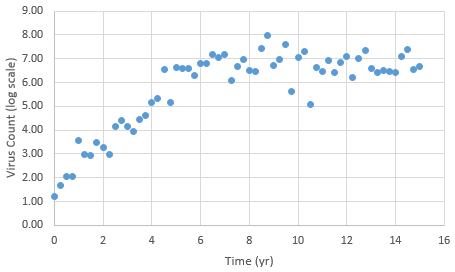
With guess values for parameters (kr1..6), approximately match the laboratory data for this patient as an initial solution. Use this initial solution to compute an optimal solution with dynamic estimation. Adjust parameters kr1..6 to match the virus count data. Start with different kr values to verify that the solution is not just locally optimal but also globally optimal.
Solution
from gekko import GEKKO
import numpy as np
# Manually enter guesses for parameters
lkr = [3,np.log10(0.1),np.log10(2e-7),\
np.log10(0.5),np.log10(5),np.log10(100)]
# Model
m = GEKKO()
# Time
m.time = np.linspace(0,15,61)
# Parameters to estimate
lg10_kr = [m.FV(value=lkr[i]) for i in range(6)]
# Variables
kr = [m.Var() for i in range(6)]
H = m.Var(value=1e6)
I = m.Var(value=0)
V = m.Var(value=1e2)
# Variable to match with data
LV = m.CV(value=2)
# Equations
m.Equations([10**lg10_kr[i]==kr[i] for i in range(6)])
m.Equations([H.dt() == kr[0] - kr[1]*H - kr[2]*H*V,
I.dt() == kr[2]*H*V - kr[3]*I,
V.dt() == -kr[2]*H*V - kr[4]*V + kr[5]*I,
LV == m.log10(V)])
# Estimation
# Global options
m.options.IMODE = 5 #switch to estimation
m.options.TIME_SHIFT = 0 #don't timeshift on new solve
m.options.EV_TYPE = 2 #l2 norm
m.options.COLDSTART = 2
m.options.SOLVER = 1
m.options.MAX_ITER = 1000
m.solve()
for i in range(5):
lg10_kr[i].STATUS = 1 #Allow optimizer to fit these values
lg10_kr[i].DMAX = 2
lg10_kr[i].LOWER = -10
lg10_kr[i].UPPER = 10
# patient virus count data
data = np.array([[0,1.20E+00],[0.25,1.67E+00],[0.5,2.06E+00],\
[0.75,2.05E+00],[1,3.57E+00],[1.25,2.96E+00],\
[1.5,2.95E+00],[1.75,3.48E+00],[2,3.27E+00], \
[2.25,2.98E+00],[2.5,4.17E+00],[2.75,4.41E+00],\
[3,4.16E+00],[3.25,3.94E+00],[3.5,4.44E+00],\
[3.75,4.60E+00],[4,5.15E+00],[4.25,5.34E+00],\
[4.5,6.56E+00],[4.75,5.16E+00],[5,6.63E+00],\
[5.25,6.60E+00],[5.5,6.59E+00],[5.75,6.28E+00],\
[6,6.79E+00],[6.25,6.81E+00],[6.5,7.16E+00],\
[6.75,7.06E+00],[7,7.19E+00],[7.25,6.07E+00],\
[7.5,6.67E+00],[7.75,6.97E+00],[8,6.51E+00],\
[8.25,6.48E+00],[8.5,7.44E+00],[8.75,7.98E+00],\
[9,6.71E+00],[9.25,6.98E+00],[9.5,7.60E+00],\
[9.75,5.62E+00],[10,7.04E+00],[10.25,7.31E+00],\
[10.5,5.08E+00],[10.75,6.62E+00],[11,6.48E+00],\
[11.25,6.91E+00],[11.5,6.44E+00],[11.75,6.85E+00],\
[12,7.09E+00],[12.25,6.20E+00],[12.5,7.02E+00],\
[12.75,7.34E+00],[13,6.57E+00],[13.25,6.42E+00],\
[13.5,6.50E+00],[13.75,6.46E+00],[14,6.42E+00],\
[14.25,7.09E+00],[14.5,7.37E+00],[14.75,6.56E+00],\
[15,6.69E+00]])
# Convert log-scaled data for plotting
log_v = data[:,1] # 2nd column of data
v = np.power(10,log_v)
LV.FSTATUS = 1 #receive measurements to fit
LV.STATUS = 1 #build objective function to match data and prediction
LV.value = log_v #v data
m.solve()
# Plot results
import matplotlib.pyplot as plt
plt.figure(1)
plt.semilogy(m.time,H,'b-')
plt.semilogy(m.time,I,'g:')
plt.semilogy(m.time,V,'r--')
plt.semilogy(data[:,][:,0],v,'ro')
plt.xlabel('Time (yr)')
plt.ylabel('States (log scale)')
plt.legend(['H','I','V'])
plt.show()
References
- Safdarnejad, S.M., Hedengren, J.D., Lewis, N.R., Haseltine, E., Initialization Strategies for Optimization of Dynamic Systems, Computers and Chemical Engineering, 2015, Vol. 78, pp. 39-50, DOI: 10.1016/j.compchemeng.2015.04.016. Article
- Hedengren, J. D. and Asgharzadeh Shishavan, R., Powell, K.M., and Edgar, T.F., Nonlinear Modeling, Estimation and Predictive Control in APMonitor, Computers and Chemical Engineering, Volume 70, pg. 133–148, 2014. Article
- Nowak, M. and May, R. M. Virus dynamics: mathematical principles of immunology and virology: mathematical principles of immunology and virology. Oxford university press, 2000.
- Lewis, N.R., Hedengren, J.D., Haseltine, E.L., Hybrid Dynamic Optimization Methods for Systems Biology with Efficient Sensitivities, Special Issue on Algorithms and Applications in Dynamic Optimization, Processes, 2015, 3(3), 701-729; doi:10.3390/pr3030701. Article
