Regression Statistics with Python
Main.PythonRegressionStatistics History
Hide minor edits - Show changes to markup
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'http://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'http://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'http://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt'
url = 'http://apmonitor.com/che263/uploads/Main/stats_data.txt'
(:html:)
<div id="disqus_thread"></div>
<script type="text/javascript">
/* * * CONFIGURATION VARIABLES: EDIT BEFORE PASTING INTO YOUR WEBPAGE * * */
var disqus_shortname = 'apmonitor'; // required: replace example with your forum shortname
/* * * DON'T EDIT BELOW THIS LINE * * */
(function() {
var dsq = document.createElement('script'); dsq.type = 'text/javascript'; dsq.async = true;
dsq.src = 'https://' + disqus_shortname + '.disqus.com/embed.js';
(document.getElementsByTagName('head')[0] || document.getElementsByTagName('body')[0]).appendChild(dsq);
})();
</script>
<noscript>Please enable JavaScript to view the <a href="https://disqus.com/?ref_noscript">comments powered by Disqus.</a></noscript>
<a href="https://disqus.com" class="dsq-brlink">comments powered by <span class="logo-disqus">Disqus</span></a>
(:htmlend:)
import pip
pip.main(['install','uncertainties'])
try:
from pip import main as pipmain
except:
from pip._internal import main as pipmain
pipmain(['install','uncertainties'])
import pip
pip.main(['install','uncertainties'])
try:
from pip import main as pipmain
except:
from pip._internal import main as pipmain
pipmain(['install','uncertainties'])
s_c = np.sqrt(pcov[2,2])
- #####################################
- Single Variable Confidence Interval
- #####################################
- #####################################
- Linear Joint Confidence Interval
- thanks to Trent Okeson
- #####################################
eigenval, eigenvec = np.linalg.eig(pcov) max_eigenval = np.argmax(eigenval) largest_eigenvec = eigenvec[:, max_eigenval] largest_eigenval = eigenval[max_eigenval]
if max_eigenval == 1:
smallest_eigenval = eigenval[0]
smallest_eigenvec = eigenvec[:, 0]
else:
smallest_eigenval = eigenval[1]
smallest_eigenvec = eigenvec[:, 1]
angle = np.arctan2(largest_eigenvec[1], largest_eigenvec[0]) if angle < 0:
angle += 2*np.pi
chisquare_val = 2.4477 theta_grid = np.linspace(0, 2*np.pi) phi = angle a_rot = chisquare_val*np.sqrt(largest_eigenval) b_rot = chisquare_val*np.sqrt(smallest_eigenval)
ellipse_x_r = a_rot*np.cos(theta_grid) ellipse_y_r = b_rot*np.sin(theta_grid)
R =
r_ellipse = np.stack((ellipse_x_r, ellipse_y_r)).T.dot(R) final_ellipse = r_ellipse + popt[None, 0:2]
- #####################################
- Nonlinear Joint Confidence Interval
- #####################################
m = 400
i1 = np.linspace(a3L,a3U*2,m)
m = 200
i1 = np.linspace(a3L,a3U*2.0,m)
plt.xlabel('a') plt.ylabel('b')
plt.xlabel('Slope (a)') plt.ylabel('Intercept (b)')
plt.plot(final_ellipse[:, 0], final_ellipse[:, 1], 'k-', label='Linear Joint Confidence Region')
LineWidth=3,label='Joint Confidence Region')
LineWidth=4, label='Nonlinear Joint Confidence Region')
plt.title('Parameter Statistics (y = a exp(b*x) + c)')
plt.title('Parameter Statistics (y = a x + b)')
- #####################################
- Single Variable Confidence Interval
- #####################################
- #####################################
- Linear Joint Confidence Interval
- thanks to Trent Okeson
- #####################################
eigenval, eigenvec = np.linalg.eig(pcov) max_eigenval = np.argmax(eigenval) largest_eigenvec = eigenvec[:, max_eigenval] largest_eigenval = eigenval[max_eigenval]
if max_eigenval == 1:
smallest_eigenval = eigenval[0]
smallest_eigenvec = eigenvec[:, 0]
else:
smallest_eigenval = eigenval[1]
smallest_eigenvec = eigenvec[:, 1]
angle = np.arctan2(largest_eigenvec[1], largest_eigenvec[0]) if angle < 0:
angle += 2*np.pi
chisquare_val = 2.4477 theta_grid = np.linspace(0, 2*np.pi) phi = angle a_rot = chisquare_val*np.sqrt(largest_eigenval) b_rot = chisquare_val*np.sqrt(smallest_eigenval)
ellipse_x_r = a_rot*np.cos(theta_grid) ellipse_y_r = b_rot*np.sin(theta_grid)
R =
r_ellipse = np.stack((ellipse_x_r, ellipse_y_r)).T.dot(R) final_ellipse = r_ellipse + popt[None, :]
- #####################################
- Nonlinear Joint Confidence Interval
- #####################################
plt.plot(final_ellipse[:, 0], final_ellipse[:, 1], 'k--', label='Linear Joint Confidence Region')
LineWidth=3,label='Joint Confidence Region')
LineWidth=4, label='Nonlinear Joint Confidence Region')
(:html:) <iframe width="560" height="315" src="https://www.youtube.com/embed/qOV8QaOzWXA" frameborder="0" allow="autoplay; encrypted-media" allowfullscreen></iframe> (:htmlend:)
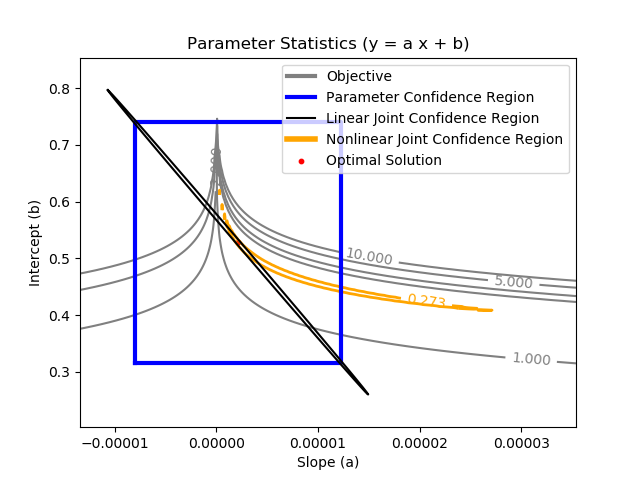
Joint Confidence Regions
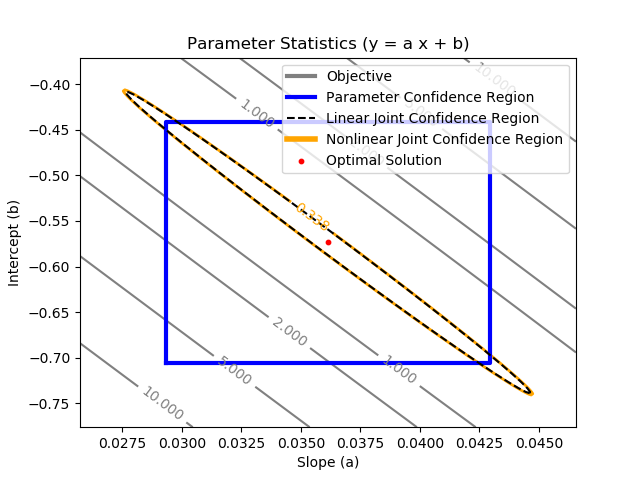
Joint Confidence Region - Linear Regression
Joint Confidence Region - Nonlinear Regression
(:toggle hide conf_int_nonlinear button show="Joint Confidence Solution":) (:div id=conf_int_nonlinear:) (:source lang=python:) import numpy as np import scipy as sp from scipy import stats from scipy.optimize import curve_fit import matplotlib.pyplot as plt import pandas as pd
- import data
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt' data = pd.read_csv(url) x = data['x'].values y = data['y'].values
def f(x, a, b, c):
return a * np.exp(b*x) + c
- perform regression
popt, pcov = curve_fit(f, x, y)
- retrieve parameter values
a = popt[0] b = popt[1] c = popt[2]
- calculate parameter stdev
s_a = np.sqrt(pcov[0,0]) s_b = np.sqrt(pcov[1,1]) s_c = np.sqrt(pcov[2,2])
- calculate 95% confidence interval
aL = a-1.96*s_a aU = a+1.96*s_a bL = b-1.96*s_b bU = b+1.96*s_b
- calculate 3sigma interval
a3L = a-3.0*s_a a3U = a+3.0*s_a b3L = b-3.0*s_b b3U = b+3.0*s_b
- Generate contour plot of SSE ratio vs. Parameters
- meshgrid is +/- change in the objective value
m = 400
i1 = np.linspace(a3L,a3U*2,m) i2 = np.linspace(b3L,b3U,m) a_grid, b_grid = np.meshgrid(i1, i2) n = len(y) # number of data points p = 2 # number of parameters
- sum of squared errors
sse = np.empty((m,m)) for i in range(m):
for j in range(m):
at = a_grid[i,j]
bt = b_grid[i,j]
sse[i,j] = np.sum((y-f(x,at,bt,c))**2)
- normalize to the optimal solution
best_sse = np.sum((y-f(x,a,b,c))**2) fsse = (sse - best_sse) / best_sse
- compute f-statistic for the f-test
alpha = 0.05 # alpha, confidence
# alpha=0.05 is 95% confidence
fstat = sp.stats.f.isf(alpha,p,(n-p)) flim = fstat * p / (n-p) obj_lim = flim * best_sse + best_sse
- Create a contour plot
plt.figure() CS = plt.contour(a_grid,b_grid,sse,[1,2,5,10], colors='gray') plt.clabel(CS, inline=1, fontsize=10) plt.xlabel('a') plt.ylabel('b')
- solid line to show confidence region
CS = plt.contour(a_grid,b_grid,sse,[obj_lim], colors='orange',linewidths=[2.0]) plt.clabel(CS, inline=1, fontsize=10)
- 2 dummy plots (gray,orange) for legend
plt.plot([aL,aL],[bL,bL],c='gray', LineWidth=3,label='Objective') plt.plot([aL,aU,aU,aL,aL],[bL,bL,bU,bU,bL],c='blue', LineWidth=3,label='Parameter Confidence Region') plt.plot([aL,aL],[bL,bL],c='orange', LineWidth=3,label='Joint Confidence Region') plt.scatter(a,b,s=10,c='red',marker='o', label='Optimal Solution')
plt.title('Parameter Statistics (y = a exp(b*x) + c)') plt.legend(loc=1)
- Save the figure as a PNG
plt.savefig('contour.png') plt.show() (:sourceend:) (:divend:)
(:toggle hide conf_int button show="Nonlinear Regression Solution":)
(:toggle hide conf_int button show="Joint Confidence Solution":)
x = data['x'].values[0:20] y = data['y'].values[0:20]
x = data['x'].values y = data['y'].values
CS = plt.contour(a_grid,b_grid,sse,[5,20,50,100,200,500],\
CS = plt.contour(a_grid,b_grid,sse,[1,2,5,10],\
(:toggle hide nonlinear button show="Nonlinear Regression Solution":) (:div id=nonlinear:)
(:toggle hide conf_int button show="Nonlinear Regression Solution":) (:div id=conf_int:)
(:toggle hide nonlinear button show="Nonlinear Regression Solution":) (:div id=nonlinear:) (:source lang=python:) import numpy as np import scipy as sp from scipy import stats from scipy.optimize import curve_fit import matplotlib.pyplot as plt import pandas as pd
- import data
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt' data = pd.read_csv(url) x = data['x'].values[0:20] y = data['y'].values[0:20]
def f(x, a, b):
return a * x + b
- perform regression
popt, pcov = curve_fit(f, x, y)
- retrieve parameter values
a = popt[0] b = popt[1]
- calculate parameter stdev
s_a = np.sqrt(pcov[0,0]) s_b = np.sqrt(pcov[1,1])
- calculate 95% confidence interval
aL = a-1.96*s_a aU = a+1.96*s_a bL = b-1.96*s_b bU = b+1.96*s_b
- calculate 3sigma interval
a3L = a-3.0*s_a a3U = a+3.0*s_a b3L = b-3.0*s_b b3U = b+3.0*s_b
- Generate contour plot of SSE ratio vs. Parameters
- meshgrid is +/- change in the objective value
m = 200
i1 = np.linspace(a3L,a3U,m) i2 = np.linspace(b3L,b3U,m) a_grid, b_grid = np.meshgrid(i1, i2) n = len(y) # number of data points p = 2 # number of parameters
- sum of squared errors
sse = np.empty((m,m)) for i in range(m):
for j in range(m):
at = a_grid[i,j]
bt = b_grid[i,j]
sse[i,j] = np.sum((y-f(x,at,bt))**2)
- normalize to the optimal solution
best_sse = np.sum((y-f(x,a,b))**2) fsse = (sse - best_sse) / best_sse
- compute f-statistic for the f-test
alpha = 0.05 # alpha, confidence
# alpha=0.05 is 95% confidence
fstat = sp.stats.f.isf(alpha,p,(n-p)) flim = fstat * p / (n-p) obj_lim = flim * best_sse + best_sse
- Create a contour plot
plt.figure() CS = plt.contour(a_grid,b_grid,sse,[5,20,50,100,200,500], colors='gray') plt.clabel(CS, inline=1, fontsize=10) plt.xlabel('Slope (a)') plt.ylabel('Intercept (b)')
- solid line to show confidence region
CS = plt.contour(a_grid,b_grid,sse,[obj_lim], colors='orange',linewidths=[2.0]) plt.clabel(CS, inline=1, fontsize=10)
- 2 dummy plots (gray,orange) for legend
plt.plot([aL,aL],[bL,bL],c='gray', LineWidth=3,label='Objective') plt.plot([aL,aU,aU,aL,aL],[bL,bL,bU,bU,bL],c='blue', LineWidth=3,label='Parameter Confidence Region') plt.plot([aL,aL],[bL,bL],c='orange', LineWidth=3,label='Joint Confidence Region') plt.scatter(a,b,s=10,c='red',marker='o', label='Optimal Solution')
plt.title('Parameter Statistics (y = a x + b)') plt.legend(loc=1)
- Save the figure as a PNG
plt.savefig('contour.png') plt.show() (:sourceend:) (:divend:)
$$\frac{SSE(\theta)-SSE(\theta*)}{SSE(\theta*)}\le \frac{p}{n-p} F\left(\alpha,p,n-p\right)$$
with p=2 (number of parameters), n=100 (number of measurements), `\theta`=[a,b] (parameters), `\theta*` as the optimal parameters, SSE as the sum of squared errors, and the F statistic that has 3 arguments (alpha=confidence level, degrees of freedom 1, and degrees of freedom 2). For many problems, this creates a multi-dimensional nonlinear confidence region. In the case of 2 parameters, the nonlinear confidence region is a 2-dimensional space.
$$\frac{SSE(\theta)-SSE(\theta^*)}{SSE(\theta^*)}\le \frac{p}{n-p} F\left(\alpha,p,n-p\right)$$
with p=2 (number of parameters), n=100 (number of measurements), `\theta`=[a,b] (parameters), `\theta`* as the optimal parameters, SSE as the sum of squared errors, and the F statistic that has 3 arguments (alpha=confidence level, degrees of freedom 1, and degrees of freedom 2). For many problems, this creates a multi-dimensional nonlinear confidence region. In the case of 2 parameters, the nonlinear confidence region is a 2-dimensional space.
(:html:) <iframe width="560" height="315" src="https://www.youtube.com/embed/r4pgGD1kpYM" frameborder="0" allow="autoplay; encrypted-media" allowfullscreen></iframe> (:htmlend:)
(:toggle hide linear button show="Linear Regression Solution":) (:div id=linear:)
(:toggle hide nonlinear button show="Nonlinear Regression Solution":) (:div id=nonlinear:)
import uncertainties as unc
import uncertainties as unc
import uncertainties as unc
(:toggle hide linear button show="Linear Regression Solution":) (:div id=linear:) (:source lang=python:) import numpy as np from scipy.optimize import curve_fit import matplotlib.pyplot as plt from scipy import stats import pandas as pd
- pip install uncertainties, if needed
try:
import uncertainties.unumpy as unp
import uncertainties as unc
except:
import pip
pip.main(['install','uncertainties'])
import uncertainties.unumpy as unp
import uncertainties as unc
- import data
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt' data = pd.read_csv(url) x = data['x'].values y = data['y'].values n = len(y)
def f(x, a, b, c):
return a * np.exp(b*x) + c
popt, pcov = curve_fit(f, x, y)
- retrieve parameter values
a = popt[0] b = popt[1] c = popt[2] print('Optimal Values') print('a: ' + str(a)) print('b: ' + str(b)) print('c: ' + str(c))
- compute r^2
r2 = 1.0-(sum((y-f(x,a,b,c))**2)/((n-1.0)*np.var(y,ddof=1))) print('R^2: ' + str(r2))
- calculate parameter confidence interval
a,b,c = unc.correlated_values(popt, pcov) print('Uncertainty') print('a: ' + str(a)) print('b: ' + str(b)) print('c: ' + str(c))
- plot data
plt.scatter(x, y, s=3, label='Data')
- calculate regression confidence interval
px = np.linspace(14, 24, 100) py = a*unp.exp(b*px)+c nom = unp.nominal_values(py) std = unp.std_devs(py)
def predband(x, xd, yd, p, func, conf=0.95):
# x = requested points
# xd = x data
# yd = y data
# p = parameters
# func = function name
alpha = 1.0 - conf # significance
N = xd.size # data sample size
var_n = len(p) # number of parameters
# Quantile of Student's t distribution for p=(1-alpha/2)
q = stats.t.ppf(1.0 - alpha / 2.0, N - var_n)
# Stdev of an individual measurement
se = np.sqrt(1. / (N - var_n) * np.sum((yd - func(xd, *p)) ** 2))
# Auxiliary definitions
sx = (x - xd.mean()) ** 2
sxd = np.sum((xd - xd.mean()) ** 2)
# Predicted values (best-fit model)
yp = func(x, *p)
# Prediction band
dy = q * se * np.sqrt(1.0+ (1.0/N) + (sx/sxd))
# Upper & lower prediction bands.
lpb, upb = yp - dy, yp + dy
return lpb, upb
lpb, upb = predband(px, x, y, popt, f, conf=0.95)
- plot the regression
plt.plot(px, nom, c='black', label='y=a exp(b x) + c')
- uncertainty lines (95% confidence)
plt.plot(px, nom - 1.96 * std, c='orange', label='95% Confidence Region') plt.plot(px, nom + 1.96 * std, c='orange')
- prediction band (95% confidence)
plt.plot(px, lpb, 'k--',label='95% Prediction Band') plt.plot(px, upb, 'k--') plt.ylabel('y') plt.xlabel('x') plt.legend(loc='best')
- save and show figure
plt.savefig('regression.png') plt.show() (:sourceend:) (:divend:)
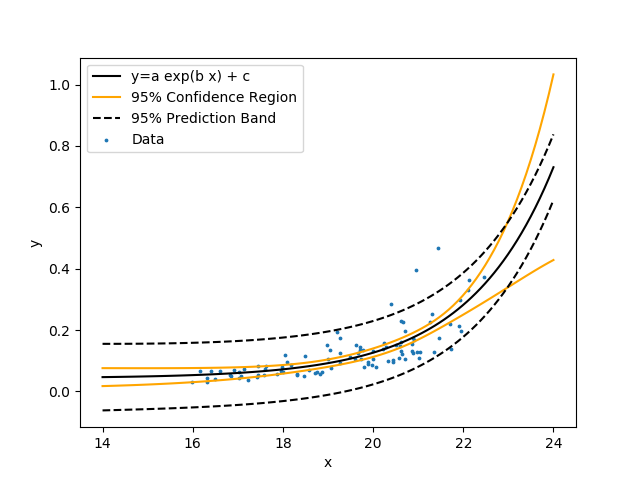
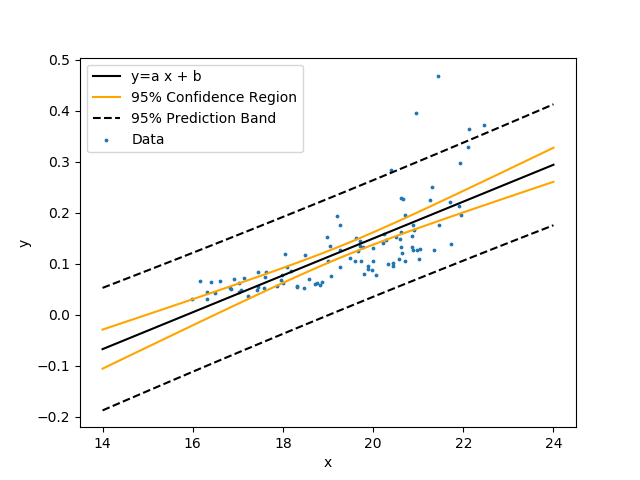
Data regression is adjusts parameter values to best fit the data. Parameter uncertainty and the predicted outcome uncertainty is important for qualifying the degree of confidence in the solution. This tutorial shows how to perform a statistical analysis with Python.
Regression is an optimization method for adjusting parameter values so that a correlation best fits data. Parameter uncertainty and the predicted uncertainty is important for qualifying the confidence in the solution. This tutorial shows how to perform a statistical analysis with Python for both linear and nonlinear regression.
plt.scatter(x, y, s=3)
plt.scatter(x, y, s=3, label='Data')
plt.plot(px, nom, c='black')
plt.plot(px, nom, c='black', label='y=a x + b')
plt.plot(px, nom - 1.96 * std, c='orange')
plt.plot(px, nom - 1.96 * std, c='orange', label='95% Confidence Region')
plt.plot(px, lpb, 'k--')
plt.plot(px, lpb, 'k--',label='95% Prediction Band')
plt.legend(loc='best')
data = pd.read_csv(url+file)
data = pd.read_csv(url)
import matplotlib.pyplot as plt import uncertainties.unumpy as unp
import matplotlib.pyplot as plt
- pip install uncertainties, if needed
try:
import uncertainties.unumpy as unp
except:
import pip
pip.main(['install','uncertainties'])
import uncertainties.unumpy as unp
data = pd.read_csv(url)
data = pd.read_csv(url+file)
plt.ylabel('y') plt.xlabel('x')
url = 'https://apmonitor.com/che263/uploads/Main/' file = 'stats_data.txt' data = pd.read_csv(url+file)
url = 'https://apmonitor.com/che263/uploads/Main/stats_data.txt' data = pd.read_csv(url)
import numpy as np from scipy.optimize import curve_fit import uncertainties as unc import matplotlib.pyplot as plt import uncertainties.unumpy as unp from scipy import stats import pandas as pd
- import data
url = 'https://apmonitor.com/che263/uploads/Main/' file = 'stats_data.txt' data = pd.read_csv(url+file) x = data['x'].values y = data['y'].values n = len(y)
def f(x, a, b):
return a * x + b
popt, pcov = curve_fit(f, x, y)
- retrieve parameter values
a = popt[0] b = popt[1] print('Optimal Values') print('a: ' + str(a)) print('b: ' + str(b))
- compute r^2
r2 = 1.0-(sum((y-f(x,a,b))**2)/((n-1.0)*np.var(y,ddof=1))) print('R^2: ' + str(r2))
- calculate parameter confidence interval
a,b = unc.correlated_values(popt, pcov) print('Uncertainty') print('a: ' + str(a)) print('b: ' + str(b))
- plot data
plt.scatter(x, y, s=3)
- calculate regression confidence interval
px = np.linspace(14, 24, 100) py = a*px+b nom = unp.nominal_values(py) std = unp.std_devs(py)
def predband(x, xd, yd, p, func, conf=0.95):
# x = requested points
# xd = x data
# yd = y data
# p = parameters
# func = function name
alpha = 1.0 - conf # significance
N = xd.size # data sample size
var_n = len(p) # number of parameters
# Quantile of Student's t distribution for p=(1-alpha/2)
q = stats.t.ppf(1.0 - alpha / 2.0, N - var_n)
# Stdev of an individual measurement
se = np.sqrt(1. / (N - var_n) * np.sum((yd - func(xd, *p)) ** 2))
# Auxiliary definitions
sx = (x - xd.mean()) ** 2
sxd = np.sum((xd - xd.mean()) ** 2)
# Predicted values (best-fit model)
yp = func(x, *p)
# Prediction band
dy = q * se * np.sqrt(1.0+ (1.0/N) + (sx/sxd))
# Upper & lower prediction bands.
lpb, upb = yp - dy, yp + dy
return lpb, upb
lpb, upb = predband(px, x, y, popt, f, conf=0.95)
- plot the regression
plt.plot(px, nom, c='black')
- uncertainty lines (95% confidence)
plt.plot(px, nom - 1.96 * std, c='orange') plt.plot(px, nom + 1.96 * std, c='orange')
- prediction band (95% confidence)
plt.plot(px, lpb, 'k--') plt.plot(px, upb, 'k--')
- save and show figure
plt.savefig('regression.png') plt.show()
Parameter Confidence
The nonlinear confidence interval is shown by producing a contour plot of the SSE objective function with variations in the two parameters. The optimal solution is shown at the center of the plot and the objective function becomes worse (higher) away from the optimal solution.
Joint Confidence Regions
The joint confidence region is shown by producing a contour plot of the SSE objective function with variations in the two parameters. The optimal solution is shown at the center of the plot and the objective function becomes worse (higher) away from the optimal solution.
$$y = a \exp{b\, x} + c$$
$$y = a \exp{\left(b\, x\right)} + c$$
$$\frac{SSE(\theta)-SSE(\theta^*)}{SSE(\theta^*)}\le \frac{p}{n-p} F\left(\alpha,p,n-p\right)$$
with p=2 (number of parameters), n=100 (number of measurements), `\theta`=[a,b] (parameters), `\theta^*` as the optimal parameters, SSE as the sum of squared errors, and the F statistic that has 3 arguments (alpha=confidence level, degrees of freedom 1, and degrees of freedom 2). For many problems, this creates a multi-dimensional nonlinear confidence region. In the case of 2 parameters, the nonlinear confidence region is a 2-dimensional space.
$$\frac{SSE(\theta)-SSE(\theta*)}{SSE(\theta*)}\le \frac{p}{n-p} F\left(\alpha,p,n-p\right)$$
with p=2 (number of parameters), n=100 (number of measurements), `\theta`=[a,b] (parameters), `\theta*` as the optimal parameters, SSE as the sum of squared errors, and the F statistic that has 3 arguments (alpha=confidence level, degrees of freedom 1, and degrees of freedom 2). For many problems, this creates a multi-dimensional nonlinear confidence region. In the case of 2 parameters, the nonlinear confidence region is a 2-dimensional space.
Use the data at the link below to create a linear and nonlinear regression.
Create a linear model with unknown coefficients a (slope) and b (intercept). Fit the model to the data by minimizing the sum of squared errors between the predicted and measured y values.
$$y = a \, x + b$$
Show the linear regression with 95% confidence bands and 95% prediction bands. The confidence band is the confidence region for the correlation equation. The prediction band is the region that contains approximately 95% of the measurements. If another measurement is taken, there is a 95% chance that it falls within the prediction band. Use the following data set that is a table of comma separated values.
(:toggle hide stats button show="Python Solution":) (:div id=stats:)
(:toggle hide linear button show="Linear Regression Solution":) (:div id=linear:)
Nonlinear Parameter Confidence Region
Nonlinear Regression
Create a nonlinear regression with unknown coefficients a, b, and c.
$$y = a \exp{b\, x} + c$$
Similar to the linear regression, fit the model to the data by minimizing the sum of squared errors between the predicted and measured y values. Show the nonlinear regression with 95% confidence bands and 95% prediction bands.
Parameter Confidence
Nonlinear Parameter Confidence Region
The nonlinear confidence interval is shown by producing a contour plot of the SSE objective function with variations in the two parameters. The optimal solution is shown at the center of the plot and the objective function becomes worse (higher) away from the optimal solution.
The parameter confidence region is determined with an F-test that specifies an upper limit to the deviation from the optimal solution
$$\frac{SSE(\theta)-SSE(\theta^*)}{SSE(\theta^*)}\le \frac{p}{n-p} F\left(\alpha,p,n-p\right)$$
with p=2 (number of parameters), n=100 (number of measurements), `\theta`=[a,b] (parameters), `\theta^*` as the optimal parameters, SSE as the sum of squared errors, and the F statistic that has 3 arguments (alpha=confidence level, degrees of freedom 1, and degrees of freedom 2). For many problems, this creates a multi-dimensional nonlinear confidence region. In the case of 2 parameters, the nonlinear confidence region is a 2-dimensional space.
(:title Regression Statistics with Python:) (:keywords data regression, uncertainty, Python, unumpy, linear regression, prediction band, confidence interval:) (:description Statistics for confidence interval and prediction band from a linear or nonlinear regression. The uncertainties package is used in Python to generate the confidence intervals.:)
Data regression is adjusts parameter values to best fit the data. Parameter uncertainty and the predicted outcome uncertainty is important for qualifying the degree of confidence in the solution. This tutorial shows how to perform a statistical analysis with Python.
Use the data at the link below to create a linear and nonlinear regression.
(:toggle hide stats button show="Python Solution":) (:div id=stats:) (:source lang=python:)
(:sourceend:) (:divend:)
Linear and nonlinear regression tutorials can also available in Python, Excel, and Matlab. A multivariate nonlinear regression case with multiple factors is available with example data for energy prices in Python. Click on the appropriate link for additional information.
(:html:)
<div id="disqus_thread"></div>
<script type="text/javascript">
/* * * CONFIGURATION VARIABLES: EDIT BEFORE PASTING INTO YOUR WEBPAGE * * */
var disqus_shortname = 'apmonitor'; // required: replace example with your forum shortname
/* * * DON'T EDIT BELOW THIS LINE * * */
(function() {
var dsq = document.createElement('script'); dsq.type = 'text/javascript'; dsq.async = true;
dsq.src = 'https://' + disqus_shortname + '.disqus.com/embed.js';
(document.getElementsByTagName('head')[0] || document.getElementsByTagName('body')[0]).appendChild(dsq);
})();
</script>
<noscript>Please enable JavaScript to view the <a href="https://disqus.com/?ref_noscript">comments powered by Disqus.</a></noscript>
<a href="https://disqus.com" class="dsq-brlink">comments powered by <span class="logo-disqus">Disqus</span></a>
(:htmlend:)